Genome Beacons - Beacon Protocol Documentation¶
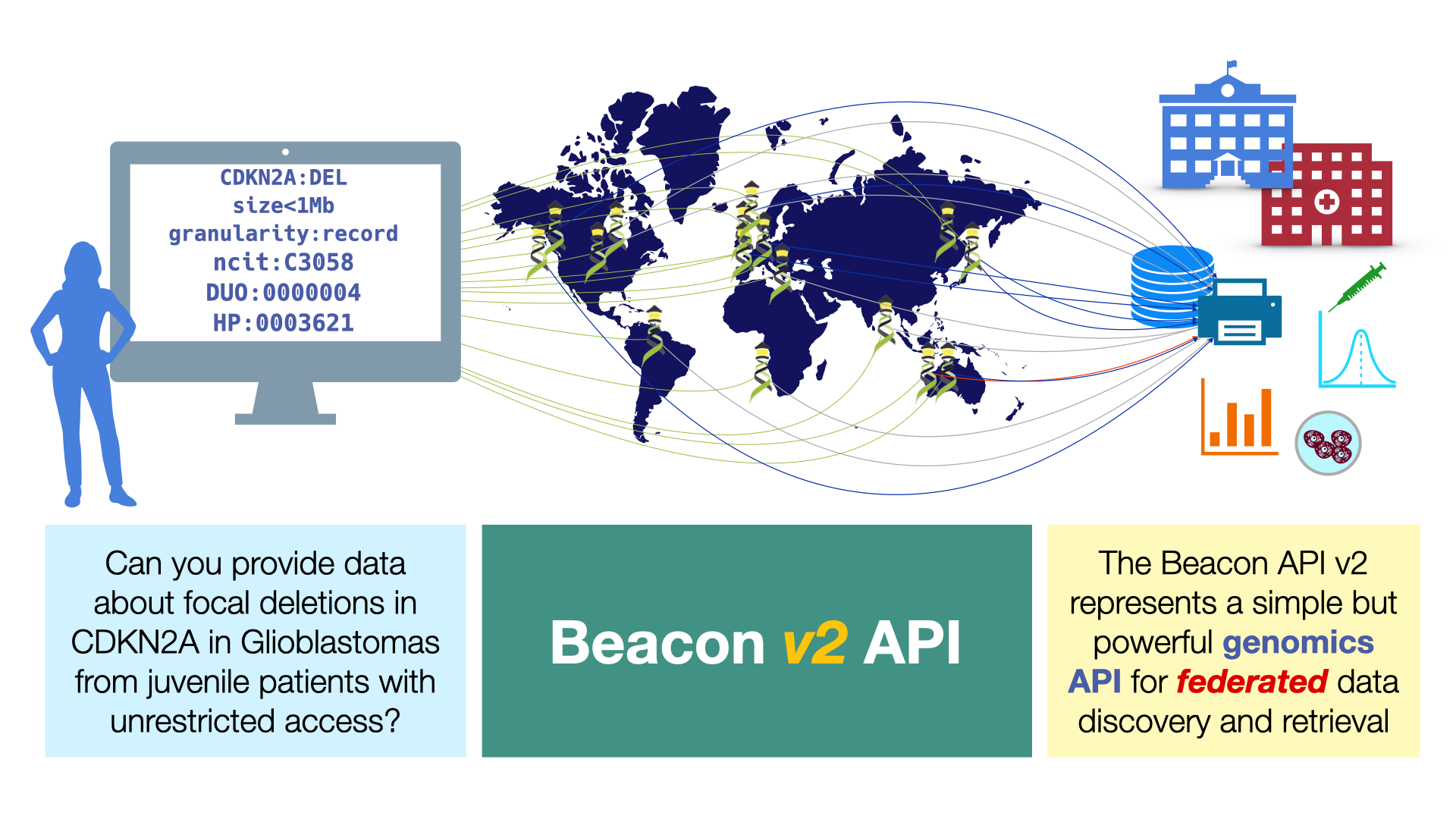
Beacon is a protocol and specification established by the Global Alliance for Genomics and Health (GA4GH). It defines an open standard for the discovery of genomic variants and biomedical data in single or distributed resources. It empowers federated data models, i.e. the discovery - and potential retrieval - of data from different organisational and geographic locations using shared standards.
The Beacon specification is developed by an international team of sientists and technology experts, as a product of the GA4GH Discovery work stream and with major support from the European bioinformatics infrastructure organization ELIXIR.
The current v2 version of the Beacon protocol has number of powerful features:
- genomic variation queries, including region queries (i.e. returning variants within a genomic/chromosomal region)
- support for information about samples or subjects (e.g. phenotypes or
other biomedical parameters), depending on the potentially required authentication and authorization
- feature queries are powered by filters with options to control the use of hierarchical terms and the precision of term matching
- data delivery using the Beacon data model or through handover
protocols
- handovers allows for data in different formats - such as VCF for genomic variants or Phenopackets - and the use of additional authentication options
Move to Beacon v2!
On 2022-04-21 Beacon v2 has been approved as an official GA4GH standard through the GA4GH steering committee.
Beacon v1 and earlier are not longer supported. Deployers of Beacon instances or networks should migrate to v2 of the standard. The functionality of Beacon v1 can be easily implemented in v2.
This website represents mostly technical information about the Beacon framework and data model and about ways towards to "beaconize" genomics datasets and resources. Additional information about the Beacon project - including news, events, publications - is available through genomebeacons.org.
Historical Tip
Originally, the Beacon protocol (versions 0 and 1) allowed researchers only to get information about the presence/absence of a given, specific, genomic mutation in a set of data but did not support phenotypic or other query parameters or the retrieval of matched records.
Components¶
Beacon consists of two components, the Framework and the Models. The Framework contains the format for the requests and responses, whereas the Models define the structure of the biological data response. The overall function of these components is to provide the instructions to design a REST API (REpresentational State Transfer Application Programming Interface) e.g. using a Beacon's OpenAPI specification (OAS).
Framework interdependency, releases and alternative models
In principle, this dual system allows for different Models (in other domains outside of the Beacon v2 model realm, e.g. "Imaging Beacon" to be built using the same Framework. However, in the current context of Beacon v2, we consider the two elements interdependent and likely to be updated together for subsequent major versions (e.g. from v2 to v3).
Informations for Different Types of Beacon Users¶
The Beacon documentation provides information for different types of users, depending on their interests and use cases. Although those will overlap, we highlight information relevant for some general scenarios throughout the documentation.
Users¶
A Beacon user (or end-user) is interested in querying Beacon instances and networks, either through web interfaces by using the Beacon API. While users of Beacon web forms in principle do not need to understand the underlying query syntax and response formats they too may benefit from some insights into the general capabilities of the underlying protocol.
User
- Beacon Models
- Knowing what is available in an instance
- Data Models and Schemas
- Beacon Flavours & Response Granularity
- Security
- Other Request, Response & Error Elements
- Using Beacon Features
- Genomic Variant Queries
- Filters for Phenotypes, Diseases & Other Parameters
- Alternative Schemas Link
Deployers and Implementers¶
A Beacon Deployer is someone who wants to make their genomics resource accessible through the Beacon protocol, without necessarily being interested or experienced in the computational aspects; while a Beacon Implementer provides the technical expertise (and potentially may get involved with Beacon development itself, e.g. to extend the protocol for novel use cases).
Deployer
-
Beacon Models
-
Reference Implementation Link
- Infrastructure requirements
- How to install
- Configuration
- Cohorts and/or Datasets
- Entry types
- Filtering terms
- Alternative schemas
- Granularity & Security
- Administration
- Testing the instance