Overview of the JSON schema pedigree.json¶
The JSON schema for the pedigree data defines how members of a pedigree (family tree) are represented. It includes several key components:
- "id": A unique identifier for the pedigree, which is distinct from a family ID. This is because one individual can be involved in multiple pedigrees related to different diseases.
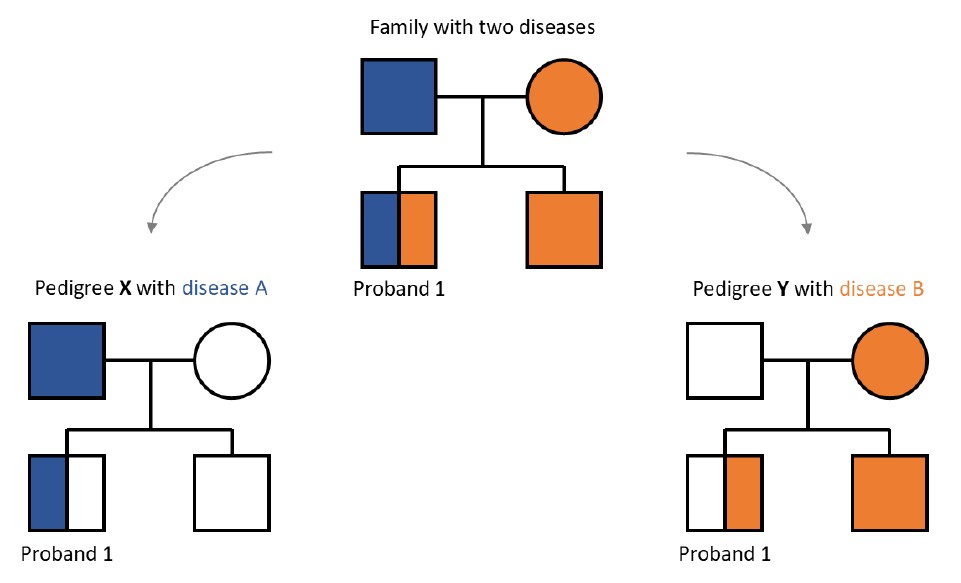
Example: in case a proband (Proband 1) is affected by two diseases segregating in the family, they can be found in two pedigrees related to these different diseases (pedigree drawn here from the Proband 1 perspective showing all family members).
-
"members": An array of individuals involved in the pedigree, either with a full family structure (if attached to the proband) or with two members (if the perspective is from a parent or relative). Each member object includes:
- "affected": Whether the individual is affected by the disease.
- "memberId": The unique identifier of the individual.
- "role": A role object identifying the relationship of the individual (e.g., proband, biological mother, biological father).
-
"disease": A reference to the disease(s) affecting the individual in the pedigree. This links to another file,
disease.json, which contains more details about the disease data. -
"numSubjects": The total number of individuals in the pedigree. This simplifies queries by providing the total count, helping to avoid reconstructing the family tree repeatedly.
Asymmetry in "members":¶
- If the schema is defined from the proband's perspective, all members (proband, parents, etc.) are included.
- If defined from another individual's perspective (e.g., the mother), only two entries are included: the individual and the proband. This structure helps with flexibility in representing different family views and cases where an individual may appear in multiple pedigrees.
FAQs and considerations:¶
1. Is it necessary to include one pedigree for each disease an individual has?
Answer: No, it is not necessary to create a new pedigree for every disease an individual has. Doing so can overburden the system with redundant information, resulting in many pedigrees that may be of little to no interest. Instead, you should focus on the most relevant or impactful diseases for the family and research context.
2. Can pedigrees be added on demand?
Answer: Yes, pedigrees can be added dynamically as the individual’s data in Beacon is not static. This allows flexibility to update an individual’s genetic or family data as more information is discovered, or when a particular disease becomes the focus of research or clinical interest.